Abstract
Background: Early and accurate outcome prediction is essential in the clinical management of high-grade B-cell lymphoma (HGBL). An early switch to salvage treatments for those patients who are likely to develop refractory/relapsed disease may improve overall survival. Unfortunately, the positive predictive value of an interim PET-CT is not high enough to guide treatment decisions. Extracellular vesicle-associated microRNAs (EV-miRNAs) are considered promising liquid biopsy-based biomarkers for lymphomas. We performed small RNA sequencing of plasma samples collected during treatment and applied machine learning to build EV-miRNA signatures for early response prediction in patients with HGBL with MYC and BCL2 and/or BCL6 rearrangements (HGBL-DH/TH).
Methods: We analyzed PAXgene ccfDNA plasma samples, from 38 of the 97 patients included in the HOVON-152 trial (NCT03620578), a prospective, multicenter, non-randomized phase II trial. In this trial, patients with HGBL-DH/TH receive one cycle of rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) followed by five cycles of dose adjusted etoposide, prednisone, vincristine, cyclophosphamide, doxorubicin, and rituximab (DA-EPOCH-R). Patients who achieve complete metabolic response (CMR) at the end-of-induction (EOI) receive one year of nivolumab consolidation. Response was assessed by an EOI PET-CT and classified as CMR (Deauville score 1-3) (responders) or no-CMR (non-responders). 20 responders and 18 non-responders (enrichment for non-responders), were selected for this analysis. We isolated plasma EVs with size exclusion chromatography as confirmed with transmission electron microscopy (TEM), tunable resistive pulse sensing (TRPS), and western blot (WB). Library preparation was done according to an unbiased, unique molecular identifier (UMI)-enhanced small RNA sequencing protocol (IsoSeek) (van Eijndhoven et al., 2021. bioRxiv) and sequenced on the NovaSeq 6000 (Illumina). We applied machine learning including the least absolute shrinkage and selection operator (LASSO), Elastic Net, and Ridge Regression to build models with EV-miRNAs for early EOI response prediction. We selected the most optimal model based on the lowest misclassification error. Then the model was internally validated and tested with bootstrapping (1000x) and over optimism estimate as well as the adjusted AUC was calculated.
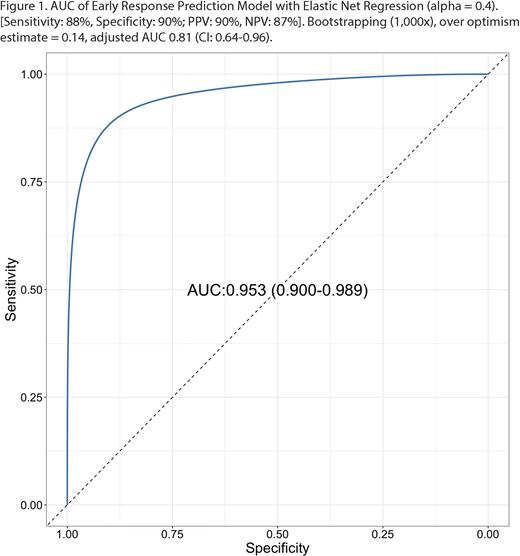
Results: TEM after one cycle of R-CHOP revealed abundant particles < 200 nm and TRPS measured significantly higher particle concentration in EV-enriched fractions. Plasma EVs were positive for CD63, CD81, flotillin-1, syntenin, HSP70, and negative for calnexin as determined by WB in accordance with the Minimal Information for Studies of EVs (MISEV) criteria 2018 for EV characterization. The most optimal model was an Elastic Net regression (a = 0.4) model consisting of 199 miRNAs with an area under the curve (AUC) of 0.95 (Confidence Interval (CI): 0.90 - 0.99) (Figure 1) [Sensitivity: 88%, Specificity: 90%; Positive Predictive Value (PPV): 90%, Negative Predictive Value (NPV): 87%]. We tested our model with bootstrapping (1,000x). After taking into account of over optimism estimate of 0.14, the adjusted AUC was 0.81 (CI: 0.64-0.96), which is higher than the performance of interim PET/CT [Sensitivity: 33-87%, Specificity: 49-94%; PPV: 20-74%, NPV: 64-95%] (Burggraaff 2019, PMID: 30141066).
Conclusion: Machine learning models using plasma EV-miRNAs prepared with the IsoSeek small RNA sequencing protocol yielded a robust signature that can predict EOI response already after one cycle of R-CHOP with a NPV of 87% and a PPV of 90%. The next step is to validate this model in a large cohort of HGBL-DH/TH cases and explore its potential in all subtypes of diffuse large B-cell lymphoma. If validated in independent cohorts, this novel approach could potentially, in combination with other modalities such as cell-free DNA and interim PET-CT, guide early risk-adapted treatment strategies in aggressive high-grade B-cell lymphomas.
Disclosures
Groenewegen:ExBiome: Current Employment. Nijland:Takeda: Research Funding; Roche: Research Funding; Genmab: Consultancy. Vergote:Beigene: Consultancy; Celgene/BMS: Consultancy; Gilead/Kite: Consultancy; Janssen: Honoraria. Kersten:Roche: Honoraria, Research Funding; Mundipharma: Research Funding; BMS/Celgene: Honoraria, Research Funding; Novartis: Honoraria; Takeda: Honoraria; Kite, a Gilead Company: Honoraria, Research Funding; Miltenyi Biotec: Honoraria; Adicet Bio: Honoraria. Chamuleau:Gilead: Research Funding; Roche: Honoraria; Novartis: Honoraria; Abbvie: Honoraria; BMS/Celgene: Honoraria, Research Funding; Genmab: Research Funding. Pegtel:Gilead: Research Funding; Takeda: Consultancy, Research Funding; Amgen: Research Funding; ExBiome: Current equity holder in private company.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal